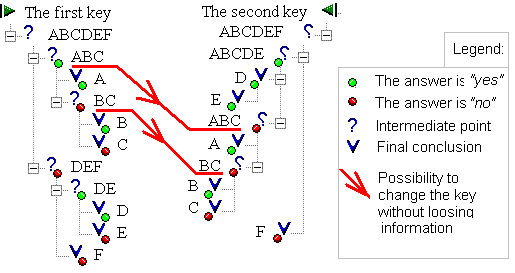
Introduction At present, a significant part of plant disease diagnostic manuals have the structure of dichotomic decision trees. Some of these manuals were written well before computers became widely available. The question arises, would this information benefit significantly from integrating into computer programs? The answer is not so obvious if we simply display the manual text on the screen, and perhaps provide additional means to navigate between questions. It is more sensible to suggest some improvements to classic algorithms, using new techniques. Our task was to create a universal, user-friendly expert system-developing shell, able to integrate the data from several manuals into one united database. This database would minimize the effects of user errors in the original manuals, and in some cases would help to find the correct conclusion even if this goal was unattainable using the manuals separately. The algorithm No matter how competent the author of the manual, the competence of the final user may not be sufficient to answer all questions in the dichotomic key. In addition, some questions might be impossible to answer before (or after) the plant disease passes a certain stage of development. The most significant shortcoming of the "dichotomic key" structure is that a failure to answer even one single question immediately collapses all process of determining. The user can only take the other manual and restart from the beginning, risking failure again. In accordance with our idea, there is no sense in rejecting already reached (but not final) conclusions each time we change the dichotomic key. There are many "internal common points", where it is possible to switch safely from key to key without losing information and without the danger of accepting the wrong conclusion. The list of final conclusions in these "common points" coincide, as it is shown in the figure below:
In this figure, two different keys have a common point ABC. In this point, it is possible to switch between keys if we fail to answer questions ABC, BC, A, B or C in the current key. Some of these "common points" may coincide with the different taxonomic groups of plant pathogen, but others do not. Computer program can be very useful in revealing "common points" easily in keys with hundreds of questions and possible conclusions. The computer program Det 3.5, created within our research group, can read the texts of tree-based keys without significant modifications. After reading, the keys are converted into a united database which can be used within a Windows interface (Fig 1):
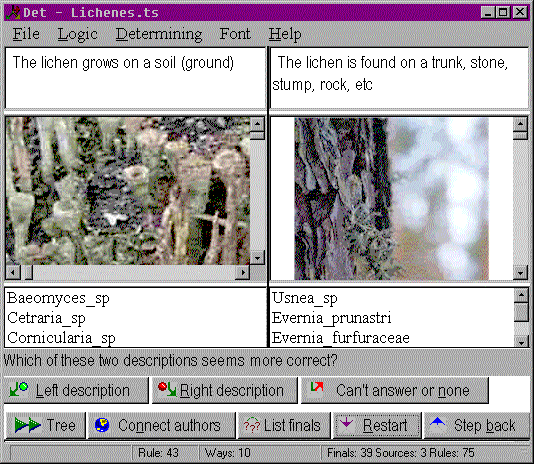
Fig 1. A typical Det session As well as the "Yes" and "No" alternatives, the user can choose the button "Can't answer nor none". In this case, the system tries to find the closest "common point" in alternative keys and switch into it. Det is also able to connect to the Internet page of the current key (if one exists). For a thorough test of the system, we wrote several keys to determine geometric figures (from the number of corners and so on). The tests showed that the system is able to progress to a correct conclusion, even if the user is unable to answer one of the questions, makes one mistake or even answers one certain category of the questions incorrectly. Hence, the system can be a serious tool in distributing professional knowledge of diagnosis of plant diseases. Det currently has about 300 users. As well as phytopathology, it is used in dermatology, criminology, polymer industry and machine diagnostic research (typical areas where such systems are needed). Det 3.5 is freeware and can be used without limitations after downloading (sites A B). Please visit the Det homepage where you will find much more information, including additional download links.End
|